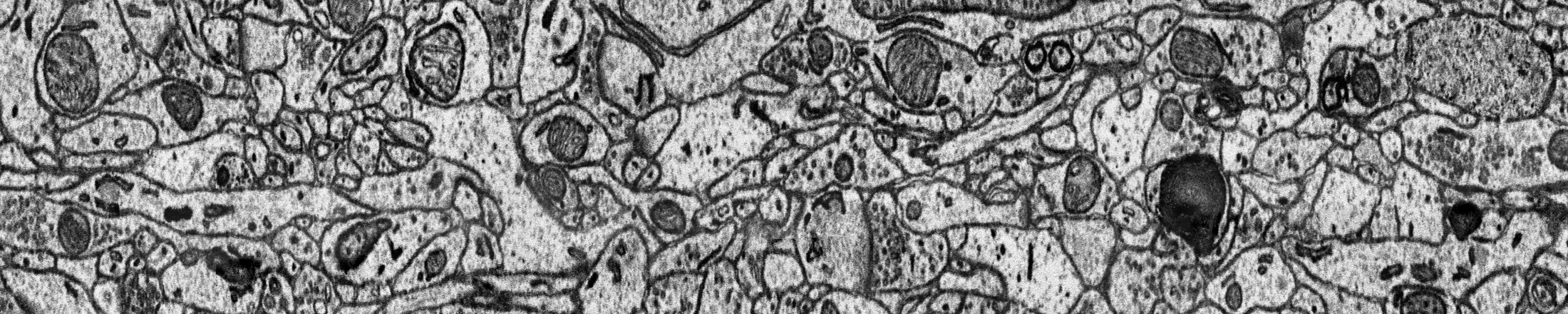
Manuscripts¶
Specific details of all algorithms submitted by participants during the ISBI challenge can be found [here].
- ngchc (VIDAR). Method: ADVANCED DEEP NETWORKS FOR 3D MITOCHONDRIA INSTANCE SEGMENTATION
- hliu (IIPPR). Method: LARGE-SCALE 3D MITOCHONDRIA INSTANCE SEGMENTATION ALGORITHM
- zhilili (VGG). Method: CONTRASTIVE LEARNING FOR MITOCHONDRIA SEGMENTATION
- cpape (EMBL). Method: AFFINITY BASED MITOCHONDRIA SEGMENTATION FOR MITOEM
- conradry71 (CEM-PDL). Method: A FAST 2D METHOD FOR INSTANCE SEGMENTATION OF DENSELY PACKED MITOCHONDRIA IN VOLUME EM DATA
- martilj (FCI). Method: AUTOMATIC INSTANCE SEGMENTATION OF MITOCHONDRIA IN ELECTRON MICROSCOPY DATA
- yliu7366 (ABCS). Method: APPLYING 3D U-NET ARCHITECTURE TO THE CHALLENGING TASK OF 3D MITOCHONDRIA SEGMENTATION IN EM IMAGES
- hiper1.1.2.3.5 (H2RNet). Method: DEEP HYBRID INSTANCE SEGMENTATION